RNA extraction-the rule of success is open!

The topic of this special article is to discuss with you the extraction of ribonucleic acid (RNA). You may have doubts here: Is RNA extraction very simple? Just follow the standard steps and add one more reagent to complete Now, is it necessary to spend a lot of time to discuss this matter? In fact, the simpler things are, the harder they are to understand. If you look at them, please be patient and listen to us.
Lab experience and customer problems
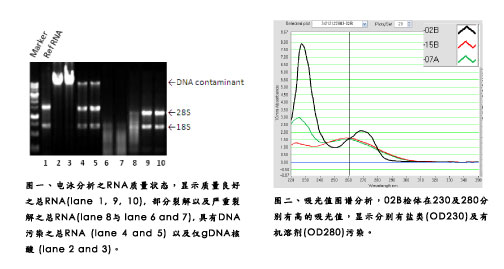
Taking Hualian Genomics Laboratory's long-term experience in accepting customers' RNA samples, the problems we often encounter are (1) the total amount of RNA is not enough, (2) RNA has serious salt pollution, (3) RNA is seriously polluted by organic solvents, (4) DNA pollution and (5) the sample is seriously degraded (Figure 1 and Figure 2). The first four problems are still easy to solve. It only needs to be extracted or purified again. However, the degradation of the sample is more difficult. Often, it takes a lot of effort and time to prepare the sample to come and go. To good results.
Customers often ask why the quality of RNA is so required to perform chip experiments. But other detection methods such as Real-time Quantitative Polymerase Chain Reaction (Q-PCR) are not needed?
Before answering this question, let ’s talk about the principle of the experiment. Simply put, the principle of Q-PCR detection is to design a pair of gene-specific primers for the gene to be detected, and cooperate with PCR through reverse transcription reaction (RT). Amplification method and timely detection of the amount of amplified product to push back the original RNA content; RT method mainly uses random primer (Random_primer) to perform complementary DNA (cDNA) replication, so even if the RNA is slightly cleaved, as Template RNA can also be faithfully turned into cDNA. However, the chip experiment is mainly based on the complementary RNA (cRNA) that is completely matched with the amplification RNA and mRNA, and then the hybridization reaction. This step must use the poly (dT) -T7 promoter sequence and the poly (A) of the signaling RNA. ) After binding, insert the T7 promoter sequence into the 5 end of the reverse transcribed cDNA, and then drive the reverse transcription reaction in the test tube through the T7 promoter to generate a reverse-stranded cRNA, and then perform subsequent chip hybridization experiments . It is conceivable that if the RNA is severely degraded, it means that many mRNAs cannot be faithfully amplified and therefore will produce many false negative results. At this point, the reader may have a concept in mind. Assuming that Q-PCR and chips are used in the storyline of the blind man touching the elephant, as long as the person using Q-PCR can touch the ear of the elephant, it can be said that there is an elephant. The number of elephants can be calculated by counting the number of ears; but those who use the chip experiment method must first confirm that the individual they touch has the complete characteristics of an elephant, in order to truly say that there is an elephant. This also explains why chip experiments require such quality of RNA.
What are the methods of RNA extraction?
In addition to traditional purification methods, such as LiCl precipitation, etc., in response to the huge demand for RNA research, there are currently many dazzling RNA extraction kits available on the market to provide research on different extraction needs, and the design principles of these products are nothing more than Use organic solvents for layered extraction such as: (1) phenol-chloroform; or use RNA to show hydrophobic properties in alcohol for adsorption separation such as: (2) Adsorption column (Silica column). Generally speaking, as long as you have the correct RNA operation concept and correct steps, you can generally extract good quality RNA, but the RNA composition distribution obtained by different extraction methods will be different, so it is recommended to be based on the analysis For target RNA, choose the correct extraction method or set. For example, if you want to study the performance of small RNA, you cannot choose the method of extracting mRNA. In addition, for samples with high levels of endogenous ribonuclease (RNase), it is recommended to use a phenol-chloroform-containing lysate to increase the ability to remove enzyme activity. In the following, I will take the operation steps of TRIzol as an example to share with readers the matters needing attention in the RNA extraction step and the method for improving the value and amount of extracted RNA.
How to clean utensils?
In the process of RNA extraction, the biggest enemy we will face is RNase. RNase is a very tenacious RNA-degrading enzyme. It is almost ubiquitous. In our environment, there are endogenous RNases on the body surface and even in cells. RNase does not need coenzyme or cofactor to exhibit activity. Although it will lose activity in the environment of denaturing solution, it will still show some activity even after removing denatured substances, even in low concentration SDS solution. It has activity, so it can effectively eliminate RNase pollution and RNA extraction is half successful. Therefore, the first task before the operation is to dry all the instruments at 180 ℃ for 6 hours or longer; or soak them in 3% H2O2 for more than 30 minutes, and then rinse them with DEPC water, Use aluminum foil after sterilization. When performing extraction, it must be in a clean, dust-free environment. If the environment permits, it is best to operate in a biological cabinet or chemical cabinet.
How are samples collected / preserved?
Earlier we mentioned that there are endogenous RNases in cells, and their expression levels vary greatly in different types of tissues. For example, brain tissue and heart tissue have relatively low endogenous RNase, but thymus tissue and pancreas tissue retain about 100,000 times the endogenous RNase of brain tissue. Therefore, it is best to freeze the animal tissues with liquid nitrogen first, and then carry out subsequent processing. It is not recommended to directly throw them into the -80 ℃ refrigerator. Because rapid freezing can freeze the activity of RNase, but freezing at -80 ° C is considered to slow down slowly, and it will still cause the degradation of some RNA; if the specimen cannot be frozen quickly, a commercially available specimen preservation solution can also be used, which can also achieve good results. .
How to crush and homogenize the sample?
Regarding the fragmentation and homogenization of the sample, the focus of this step should be on how to allow the organic lysate to quickly infiltrate the specimen. Phenol-chloroform can dissolve the cells, which will cause protein denaturation, so the activity of endogenous RNase will go away. Activation; on the contrary, if the cells cannot be quickly lysed to expose the protein and then denature, RNase will degrade RNA, so how to quickly lyse cells of different tissues is a subject worth thinking about and adjusting, and also the key to success or failure. As far as the type of specimen is concerned, generally, the cultured cells are mostly monolayer cells, which will not encounter such a problem, and the extraction is relatively easy, but the extraction of three-dimensional tissues will encounter the problem of uneven homogenization and cause RNA degradation. Therefore, low temperature liquid nitrogen milling is the most effective way to break tissues, but liquid nitrogen milling is more troublesome. If it encounters a large number of samples, it is time-consuming and ineffective, but if conditions permit, this is still a good idea. However, considering efficiency and speed, a homogenizer is a better choice, but the process of using a homogenizer has several important points that must be noted: (1) The speed is fast, and the cell homogenization is completed before the RNase shows activity, (2 ) Avoid heat generation, so as not to degrade RNA, (3) Do not over-grind, over-grinding will interrupt the DNA and cause DNA contamination of small fragments. In addition, there is an important point that is often overlooked in the process of homogenizing lysed cells, that is, the ratio of general cells to organic lysate is often neglected. Some operators will think that if more tissue is placed, a large amount of RNA can be obtained, but there is no Realizing that too much tissue and too little lysate, the cells cannot be completely lysed and the RNase cannot be completely denatured, but the effect is counterproductive, and finally the RNA is severely degraded. Therefore, if the homogenized solution is too viscous to be layered after the reaction, the lysis solution must be added.
Centrifugal stratification of samples
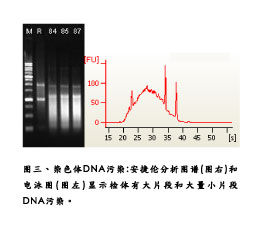
In the phenol-chloroform extraction method, the centrifugal rotation speed of the mixed solution cannot be adjusted arbitrarily, and should be fixed at 12000g rotation speed, so as not to be effectively layered due to the change of the sedimentation coefficient. After the centrifugation is completed, it can be clearly observed that the sample is divided into the upper water layer and the lower organic layer, and the RNA we want is dissolved in the upper water layer, and the volume is about 500 ~ 600μl. At this time, it is recommended to use a small caliber 100 or 200μl tip to pipette the aqueous solution into a clean 1.5ml centrifuge tube. The common mistake in this step is: the operator is eager to take out the water layer and stir the original layer; Or I feel that extraction is not easy to recover some RNA, so the water layer is taken out as much as possible, which results in the absorption of organic solution and a large amount of DNA pollution (Figure 3), causing troubles in subsequent experimental processing and even failure of the experiment, so it is strongly It is recommended to extract only 70% to 80% of the RNA aqueous solution for the next step. As the saying goes, the so-called failure must be achieved. There is no need to give up the entire forest for a tree. A timely choice can ensure that the subsequent experiment will proceed smoothly. In addition, in general, phenol and water have a certain proportion of mutual solubility, so there is still a certain amount of phenol remaining in the RNA aqueous solution at this time, it is recommended to use chloroform to perform a centrifugal delamination to remove the remaining phenol.
Precipitation of RNA samples
Generally, we will use ethanol (aqueous solution: ethanol = 1: 2.5) or isopropyl alcohol (aqueous solution: isopropyl alcohol = 1: 1) for RNA precipitation, there is no significant difference in effect, but if the amount of RNA is known to be small Alternatively, you can use the ethanol precipitation method, and place the sample in a refrigerator at -80 ℃ overnight to do centrifugation, and with the increase of the centrifugation time, this can increase the recovery rate of RNA precipitation.
RNA sample cleaning to remove salts
In order to remove the salts in the RNA precipitate, we can use 70-75% ethanol for washing. If possible, let the nucleic acid precipitate be suspended. It is best to use two washings, and then centrifuge the tube in a centrifuge. Use a pipette to carefully aspirate the residual ethanol. After a little dry or blow dry the nucleic acid precipitate to volatilize the remaining ethanol, re-dissolve with DEPC water.
Conclusion
If you have noticed and improved the parts explained above, but still can not extract good quality RNA, then maybe we must check whether our experimental design will lead to the activation of RNA degradation mechanism, for example: cells are undergoing cells The physiological reaction of apoptosis or tissue necrosis; or the treatment of the drug will directly cause RNA degradation, etc. If it is, it may be necessary to make experimental design amendments or revise the requirements for RNA quality.
The above techniques for RNA extraction are the practical experience of Hualian Genomics Laboratory in the operation of tens of thousands of various RNA samples over the years. The accumulated experience has come to some experienced readers. It may be that the savages have exposed it. However, I hope that such experience sharing can help those customers who have encountered difficulties in RNA preparation to successfully complete the preparation of RNA specimens. The laboratory takes the mission-critical spirit and takes every delivery to us seriously The sample on hand will make the most perfect result with the fastest speed and send it to the client. If the above content is not detailed or unclear, please feel free to contact Hualian Lab for discussion or experience exchange. We will definitely go all out.
Holiday Lashes,Halloween Fake Lashes,Christmas False Lashes,Costume False Lashes
Zhengzhou Cuka Electronic Commerce Co., Ltd. , https://www.cukalashes.com